|
Checking
assumptions in regression
|
|
Chong Ho Yu, Ph.D.
|
Several assumptions for the data should be met in
order to apply a valid regression model. Wonnacott and
Winacott (1981) argued that if the assumptions of
linearity, normality and independence are upheld,
additional assumptions such as fixed values of X are
not problematic. Berry & Feldman (1995) pointed
out that most regression assumptions are concerned
with residuals. This write-up will demonstrate how
different assumptions of regression are examined.
Residuals have constant variance
(homoescedasticity)
When the error term variance appears constant, the data
are considered homoscedastic, otherwise, the
data are said to be heteroscedastic. The SAS's
syntax for checking residuals are as follows:
proc reg;
model y = x1 x2;
output out=two
p=y_hat
r=y_res;
proc gplot data=two; plot y_res * y_hat;
|
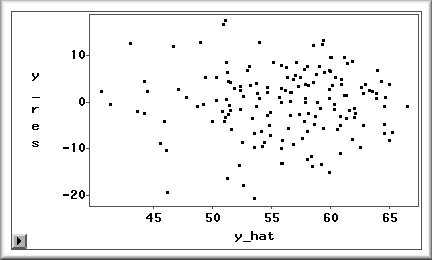
The outout is a plot of residuals versus predicted
values. If the residuals variance is around zero, it
implies that the assumption of homoscedasticity is not
violated. If there is a high concentration of
residuals above zero or below zero, the variance is
not constant and thus a systematic error exists.
Independence of Residuals
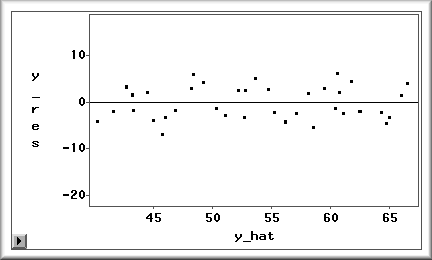
A regression model requires independence of error terms.
Again, a residuals plot can be used to check this
assumption. Random, patternless residuals imply
independent errors. Even if the residuals are even
distributed around zero and the assumption of constant
variance of residuals is satisfied, the regression model
is still questionable when there is a pattern in the
residuals as shown in the following figure.
Normality of Residuals
It is important to note that for regression the
normality test should be applied to the residuals rather
than the raw scores. There isn't a general agreement of
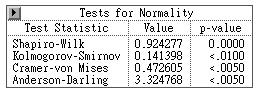
the best way to test normality. In SAS, there are four
test statistics for detecting the presence of
non-normality, namely, the Shapiro-Wilk (Shapiro &
Wilk, 1965), the Kolmogorov-Smirnov test, Cramer von
Mises test, and the Anderson-Darling test. According to
the SAS manual, if the sample size is over 2000, the
Kolmgorov test should be used. If the sample size is
less than 2000, the Shapiro test is better. The null
hypothesis of a normality test is that there is no
significant departure from normality. When the p
is more than .05, it fails to reject the null hypothesis
and thus the assumption holds.
NCSS (NCSS Statistical Software, 2007) provides more
normality tests in addition to the Shapiro test and
the Kolmgorov test (see the following table).
According to NCSS, the Shapiro and the Anderson
Darling tests are the best. The Kolmogorov test is
included just because of its historical popularity,
but is bettered in almost every way by other tests.
However, some researchers argue that the Shapiro
test was originally constructed to test a sample size
carrying up to 50 subjects. To examine normality for a
sample size between 51 to 1999, other tests such as
Anderson-Darling, Martinez-Lglewicz, and D'Agostino
tests are recommended.
Normality Test Section
|
Test
Name
|
Test
Value
|
Prob
Level
|
10%
Critical Value
|
5%
Critical Value
|
Decision
(5%)
|
|
Shapiro-Wilk W
|
0.937
|
0.21
|
|
|
Accept
Normality
|
|
Anderson-Darling
|
0.443
|
0.29
|
|
|
Accept
Normality
|
|
Martinez-Iglewicz
|
1.026
|
|
1.216
|
1.357
|
Accept
Normality
|
|
Kolmogorov-Smirnov
|
0.148
|
|
0.176
|
0.192
|
Accept
Normality
|
|
D'Agostino Skewness
|
1.037
|
0.299
|
1.645
|
1.960
|
Accept
Normality
|
|
D'Agostino Kurtosis
|
-.786
|
0.432
|
1.645
|
1.960
|
Accept
Normality
|
|
D'Agostino Omnibus
|
1.691
|
0.429
|
1.645
|
1.960
|
Accept
Normality
|
Although many authors recommended using skewness and
kurtosis for examining normality (Looney, 1995),
Wilkinson (1999) argued that skewness and kurtosis
often fail to detect distributional irregularities in
the residuals. By this argument, the D'Agostino
skewness and the D' Agostino Kurtosis test may be less
useful.
Also, statistical tests depend on sample size, and as
sample size increases, the tests often will reject
innocuous assumptions. Most normality tests have small
statistical power (probability of detecting nonnormal
data) unless the sample size is large. If the null is
rejected, the data are definitely non-normal. But if
it fails to reject the null, the conclusion is
uncertain. All you know is that there was not enough
evidence to reject the normality assumption (NCSS
Statistical Software, 2007). To rectify the sample
size problem, Hair, Anderson, Tatham and Black (1992)
suggested that in a small sample size it is safer to
use both a normal probability plot and test statistics
to ensure the normality. In SAS submitting the
following command syntax returns both normal
probability plot and test statistics.
proc univariate normal plot data=two; var y_res;
histogram y_res/normal;
QQplot y_res; run;
|
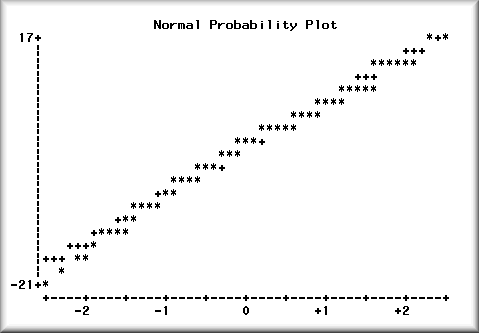
In a normal probability plot, the normal distribution
is represented by a straight line angled at 45
degrees. The standard residuals are compared against
the diagonal line to show the departure. If the
residuals follow along the straight line, it means
that the departure from normality is slight.
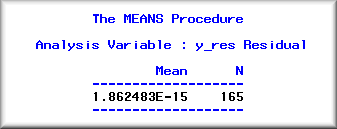
Residuals have mean as zero
The above PROC UNIVARIATE statement returns the mean.
One can also use PROC MEANS to get the same result.
However, when the mean value carries many decimals, the
SAS system will use E-notation. In the following
example, the decimal point should shift 15 positions to
the left, and thus the mean value is near zero
(.000000000000001862483).
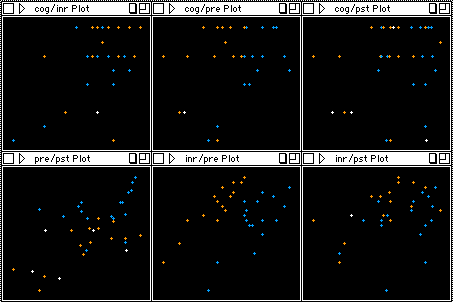
Linearity
To examine the assumption of linearity, one can apply a
scatterplot matrix showing all Xs against Y in a
pairwise manner. However, this option is not available
in SAS and SPSS's scatterplot matrix is not interactive.
I recommend using an interactive scatterplot matrix,
which is a feature of DataDesk. In a DataDesk's
scatterplot matrix, one can assign colors to the data
points for detecting clusters in different
relationships. Take the following graphs as an example,
the assumption of linearity seems to be violated because
it appears that there are two clusters within the
subjects. A linear fit to all data points is not the
best fit.
Fox (1991) suggested that although it is useful to
plot y against each x for the examination of
linearity, these plots are inadequate because they
only tell the partial relationship between y and each
x, controlling for the other xs. Therefore, it is
desirable to use residual plots against y. How to
output residuals has been illustrated in the section
of homoescedasticity. One can modify those methods by
replacing the predicted values (Y hat) with the
observed values (Y).
The absence of multicollinearity
Multicollinearity will cause the variances to be high.
These inflated variances are quite detrimental to
regression because some variables add very little or
even no new and independent information to the model
(Belsley, Kuh & Welsch, 1980). Although Schroeder,
Sjoquist and Stephen (1986) asserted that there is no
statistical test that can determine whether or not
multicollinearity really is a problem, there are still
several ways for detecting multicollinearity such as a
matrix of bivariate correlation and the regression of
each independent variable in the equation on all other
independent variables (Berry and Feldman, 1985).
Nonetheless, the former approach lacks sensitivity to
multiple correlations while the latter cannot tell much
about the influence of regressors to variances. A better
approach is using the Variance Inflation Factor (VIF).
The detail of detecting multicollinearity is in the
write-up Multicollinearity,
variance inflation factor, and orthogonalization.
Put all things together
It is more efficient to check all of the preceding
issues at the same time. The following simple SAS
macros was written for this purpose.
|
/*
name = name of the data set
dv = dependent variable,
x1 = first independent variable,
xlast = last independent variable */
%macro reg (name,
dv, x1, xlast);
proc reg data=&name; model &dv =
&x1 - &xlast/vif;
output out=two
p=y_hat
r=y_res;
title "Check multicollinearity
using VIF";
proc gplot data=two; plot y_res * y_hat;
title "Check homoescedasticity
and independence of residuals";
proc univariate normal plot data=two; var
y_res;
histogram y_res/normal;
probplot y_res;
QQplot y_res;
title "Check the mean and
normality of residuals";
proc gplot data=&name; plot &dv *
(&x1 - &xlast);
title "Check linearity";
run;
%mend reg;
/*
To invoke the macros, enter the actual
names of the data set,
DV, fist IV, and the last IV in the
following.
*/
%reg(name,
dv, x1, xlast)
|
References
- Belsley, D. A., Kuh, E. & Welsch, R. E.
(1980). Regression Diagnostics: Identifying
influential data and sources of collinearity.
John Wiley.
- Berry, W. D. & Feldman, S. (1985). Multiple
regression in practice. London: Sage
Publications.
- Fox, J. (1991). Regression diagnostics.
Sage Publications.
- Looney, S. W. (1995). How to use tests for
univariate normality to assess multivariate
normality. American Statistician, 49, 64-70.
- NCSS Statistical Software. (2007). NCSS. [Computer
Software] Kaysville, UT: Author.
- Schroeder, L. D., Sjoquist, D. L. & Stephan,
P. E. (1986). Understanding regression analysis.
Beverly Hills, CA: Sage Publications.
- Shapiro, S. S., & Wilk, M. B. (1965). An
analysis of variance test for
normality. Biometrika, 52, 591-611.
- Wilkinson, L, & Task Force on Statistical
Inference. (1999). Statistical methods in psychology
journals: Guidelines and explanations. American
Psychologist, 54, 594-604.
- Wonnacott, T. H. & Wonnacott, R. J. (1981). Regression:
A second course in statistics. Wiley.
Last updated: 2024
Navigation
Index
Simplified Navigation
Table of Contents
Search Engine
Contact
|